Former Group Neuroanatomy within Department Neurophysics – Human Microstructural Connectomics
High-Resolution Histology Informs Magnetic Resonance Imaging-Based Tractography
Recent advances in magnetic resonance imaging (MRI)-based biophysical models facilitate the estimation of white matter (WM) properties (e.g., relative myelination, axonal caliber, axonal density) in vivo. A main limitation, however, is the simplification of the microstructural tissue composition, e.g., axons are modeled as long parallel cylinders with invariable diameters. This can lead to a systematic over- or underestimation of the derived parameters. Another limitation is the scale gap between a typical MRI voxel (~ 500 µm) and the tissue microanatomy (e.g., axon diameters ~ 1µm). High-resolution light and electron microscopy images of 2-D histological data provide a more detailed understanding of the microstructural composition of WM pathways. The goal of this project is to define with this approach the microanatomical properties of WM tracts with well-characterized anatomical orientation, such as the optic chiasm, corticospinal tract, and corpus callosum. This should lead towards improved reference data for the interpretation of structural MRI findings.
We process human tissue samples obtained at autopsy with prior informed consent from subjects without neurological or psychiatric diseases, fix them with 3% paraformaldehyde and 1% glutaraldehyde, and scan them with high-resolution structural and diffusion-weighted MRI scans. We then embed the tissue in Durcupan resin and cut semithin (500 nm) and ultrathin (50 nm) sections with an ultramicrotome. Semithin sections are stained with toluidine blue and digitized with a slide scanner, ultrathin sections are imaged with a transmission electron microscope.
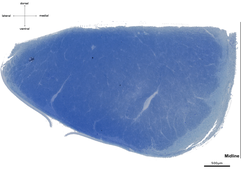
Light microscopy of semithin and especially electron microscopy of ultrathin sections are restricted by a very small field of view. To overcome this limitation, we employ a pipeline optimized for large fields of view. For a comprehensive description of tract properties we use high-resolution (250 x 250 nm in-plane) light microscopy images of cross-sections of entire tracts, e.g., corticospinal tract at the level of the medulla oblongata (~ 10 mm², figure above). Parts of these light microscopy images (~ 100 x 100 µm, figure below, left) are manually segmented into three structural compartments, (i) axonal cytoplasm (figure below, middle, green), (ii) myelin sheaths (red), and (iii) background (blue) to train a deep-learning algorithm. Structural parameters, such as axon density and diameter or myelin thickness and density can then be derived from these images. The trained deep-learning algorithm will be able to compute these microstructural parameters for the entire cross-sections of tracts (figure below, right, axonal cytoplasm in white, myelin sheaths in grey, background in black). To evaluate the quality of automatic segmentation of light microscopic images we use adjacent ultrathin sections processed for electron microscopy.

In combination with MRI data from the same tissue samples these high-resolution histological measurements should further our insight into the microstructural composition of human brain WM tracts and deepen our understanding of the morphological basis of MRI-based biophysical models.

