WP2 UvA: Computational neuroanatomy
Background
Dr. Anneke Alkemade and her team focus on integrative computational neuroanatomy research at the University of Amsterdam. The human subcortex consists of ~455 individual structures, accounting for approximately a quarter of total brain volume. Despite the well-recognized importance of the subcortex, it receives little attention compared to the neocortex. Dr. Alkemade aims to develop and apply interdisciplinary approaches to map deep brain structures, and to tackle research questions on human functional neuroanatomy. She provides resources and tools that can be applied by research groups world-wide to advance our understanding of the human subcortex using both structural and functional MRI.
Large scale lifespan qMRI dataset. Alkemade and colleagues have invested 6 years to create the Amsterdam Ultra-high field Adult Lifespan Database (AHEAD), a submillimeter 7T MRI dataset containing quantitative MRI contrasts in 105 healthy participants across the adult lifespan. This database is shared without restrictions with the scientific community and has been downloaded over 8400 times.
Automated parcellation of the human subcortex. This data was used to set up manual parcellation protocols that were applied by two independent raters to create manual parcellations of 25 individual brain structures, which can be reused for validation purposes as proposed in the current application. Manual parcellation is, however, a labor-intensive process, and is therefore not suitable for the analyses of large databases containing MRI data of many individuals. To remedy these practical limitations, the group developed the Multi-contrast Anatomical Subcortical Structure Parcellation (MASSP) algorithm. MASSP follows a Bayesian multi-object approach combining shape priors, intensity distribution models, spatial relationships, and global constraints. Currently, the MASSP algorithm has been trained successfully to parcellate 17 structures. The reliability of the MASSP algorithm for parcellations in older participants has been confirmed using manual validation in different age groups. MASSP provides a reliable parcellation framework to analyse automatically the data collected in WP1, 3 and 4.
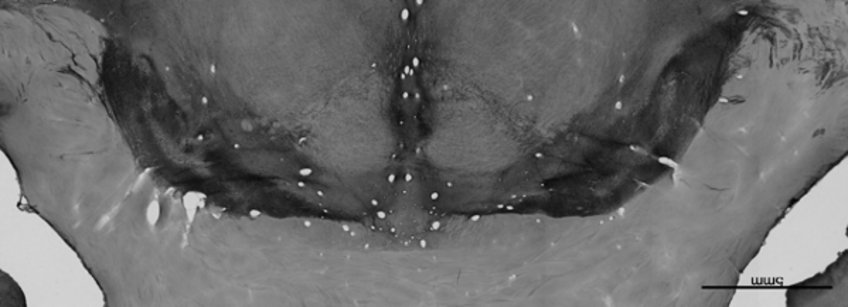
Advanced subcortex 3D histology. Still, a number of structures and substructures continue to be too small for direct visualization. To create probabilistic maps for detailed research, Alkemade and her colleagues have developed an approach that allows direct linking of histochemical data with qMRI (Fig 2B). These efforts can be reused to create probabilistic atlases of structures that at present cannot be visualized with sufficient detail in in vivo studies, or which require post mortem validation of the results such as visualizations of the nigrosomes in the current proposal. Preliminary findings on nigrosomes in SN link MRI features to underlying 3D anatomy already challenging current textbook knowledge of widely used conventional MRI-based nigrosome markers.
Resulting parcellations can readily be used to perform qMRI modeling in subcortical structures, allowing the description of iron and myelin contents, as well as shape characteristics of individual structures. Dr. Alkemade's contribution to IronSleep will allow building of detailed models of maturation and decline across the entire subcortex, and provide insight into potential pathophysiological changes in (preclinical) PD deviating from age-related variations in subcortical anatomy.
Working plan within IronSleep
Imaging of subcortical structures is challenging due to their relative distance to the MRI coil, high subcortical iron content, and the small size of individual nuclei. When the technical challenges are adequately addressed, over 25 subcortical structures can be manually parcellated using standardized protocols on submillimeter 7T qMRI. Parcellations can then be fed into the MASSP algorithm developed by our team to create automated individual parcellations over large qMRI data sets. In collaboration with Prof. Dr. Forstmann and Dr. P-L. Bazin, we will develop manual parcellation procedures for the inclusion of additional structures which are currently not included in MASSP, and which are involved in sleep-wake cycle targets. These targets include basal forebrain nuclei (nucleus basalis of Meynert, NBM), locus coeruleus (LC), and the dorsal Raphé nucleus. If sufficient contrast is present, this will be done in in vivo qMRI data. If MRI contrast does not allow reliable detection, probability maps will be created using available post mortem data (see Fig. 2B). This list of structures will be further extended with individual thalamic nuclei and hippocampal substructures which have been implicated in PD. Furthermore, post mortem and in vivo data will be used to provide more anatomical detail to allow assessment of changes in nigrosomes in the SN. MASSP will be retrained with these extensions for the analyses on MRI contrasts obtained in WP1,3 and 4, to provide the basis for modeling studies. Fine-tuning of the algorithm will be performed to seamlessly handle the diverse data sources and their resolutions, balancing prior atlas-based knowledge with individual subject data in its Bayesian framework.
Team – University of Amsterdam

Dr Anneke Alkemade

Dr Pierre-Louis Bazin
